The multiple possibilities of small microRNAs in breast cancer
Written by Dr. Vinitha Richard
While it may seem small, the ripple effect of small things is extraordinary.
Matt Bevin
The size of an early-stage primary breast tumour might range from 1mm-20mm, which could be related to the tip of a sharpened pencil (1mm) or the size of a peanut (20mm). Even a small tumour volume harbours millions of cancer cells, few of which have the ability to escape the tumour barrier even at an early stage and find their way to the bloodstream as metastatic cells. Early detection of the presence of tumours in the body increases the chances of disease-free survival.
Flooded with technological innovations and enhanced healthcare opportunities, we have surpassed several milestones in the efficient diagnosis and multiple treatment options in the fight against cancer. Over the years we have devised and implemented tactics to target the proteins and genes related to breast cancer, which is one of the easily detectable solid epithelial tumours. Yet we witness an increasing trend of high rates of breast cancer among women worldwide. This calls for readdressing the current scientific perspective of the disease and the mechanisms associated with the progression and recurrence of breast cancer in selective patients.
Just like the unique physical characteristics such as fingerprints, iris, and palm or finger vein patterns termed biometrics that can specifically recognize and identify an individual, we need a molecular biometric standard that can accurately detect the presence of cancer in an individual. Such an operational molecular biometric system should also provide information as to the response of the individual to a potential therapeutic regimen, the chances of their long-term survival, and also predict the risk of relapse after a period of disease-free status.
From the initial discovery of microRNAs in 1993 by Lee and Ambrose of Harvard University, USA, microRNAs have garnered immense attention as key players regulating gene expression right from simple roundworms to even more complex multicellular humans. Made up of a combination of four nucleotides [adenine (A), Uracil (U), Guanine (G), and cytosine(C)], microRNAs differ from the messenger RNAs (mRNAs) in the length of the chain of nucleotides (~19–24 nucleotides (nts) whereas mRNAs range from 400-1500 nts in length. MicroRNAs bind to the complementary target sequences in mRNA leading to mRNA destabilization, degradation, the resultant decrease in mRNA expression levels and thereby interfering with the final production of a functional protein product. With more than >35 000 miRNA sequences identified in >270 organisms, miRNAs play a role in crucial cell activities such as cellular signalling, metabolism, cell differentiation and proliferation, and apoptosis by controlling the expression of multiple genes and are abundant in cells.
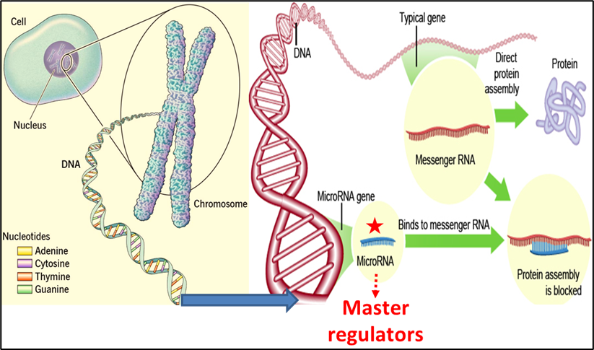
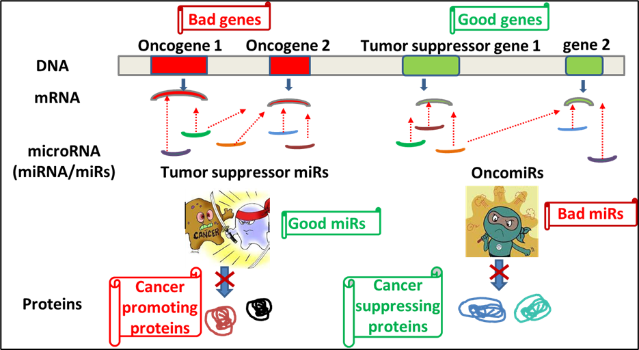
MicroRNAs may function as either oncogenes or tumour suppressors under certain conditions. ‘Oncogenic miRNAs’, are upregulated in cells and they tend to inhibit the expression of potential tumour suppressive genes, while ‘tumour suppressive miRNAs’ are down-regulated causing the augmentation of downstream signal pathways or oncogenes responsible for the development of tumours. OncomiRs and tumour suppressor miRs may target the same or different mRNAs intracellularly (tissue) or extracellularly (in circulation). Different cancers may share common aberrant miRNAs, although cancer-specific miRNAs are the major population.
Breast cancer has been categorized into various subtypes and treatment decisions have been made based on the surface expression of hormone receptor proteins oestrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor (HER2). Post-treatment and over a period of disease-free survival, several cases report the emergence of resistant cancer cells of a different subtype or receptor phenotype. This receptor discordance is attributed to the sudden loss of gain of receptor expression mediated by regulation of the receptor gene expression. MicroRNAs can modify the expression of genes post-transcriptionally and lead to the development of intratumoural heterogeneity.
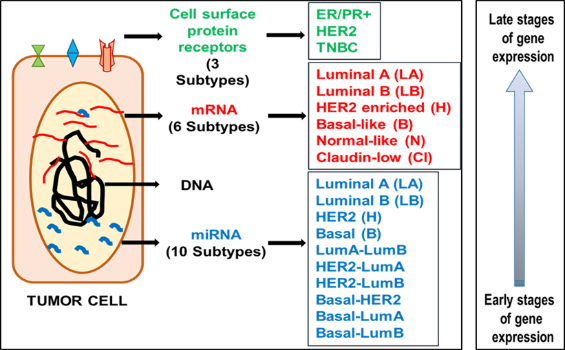
The expression profile of proteins has categorized breast cancer into three major subtypes, whereas gene expression profiling has identified 5 major groups. Further a step, an exclusive microRNA profiling of breast tumour tissue led to the identification of 10 combinations of breast tumour subtypes and this widens the angle for implementing targeted treatment or precision medicine in breast cancer. Defining each of the subtypes of breast tumours accurately will lead to the identification of the tumour cells that will succumb to treatment and those that will resist. This will enable the prediction of patients likely to respond to chemotherapy and those who will not, thus sparing them with unwanted side effects of chemo. Also elucidating the significance of the identified microRNAs will pave way for utilizing them as therapeutic targets to alter the disease status of an individual. Though they are small non-coding molecules, the ripple they create travels from cell to cell bringing about significant changes in the human body that is yet to be disclosed through research.

Dr. Vinitha Richard is a postdoctoral fellow working at the University of Galway under Prof. Michael Kerin, as part of POI work package 1.4. "MicroRNAs For The Detection Of Breast Cancer"

