P3.3: Determining the cellular and molecular heterogeneity driving acute myeloid leukaemia (AML) for the design of therapeutic strategies

Plain Language Summary
Acute myeloid leukaemia (AML) is one of the four main types of blood cancer. AML is caused by the development of abnormal form white blood cells, called myeloid cells. These abnormal myeloid cells grow uncontrollably, and they are not able to develop into a range of important immune cells. As a result, people with AML suffer from anaemia and infections.
Myeloid cells reside in the bone marrow where they interact with other types of cells. This interaction is essential for the survival of abnormal myeloid cells and it also protects some of them from chemotherapy. The AML cells sheltered from the treatment in the bone marrow can develop drug resistance. These drug-resistant cells will grow when the treatment stops and make the disease come back. Accordingly, doctors see that while AML patients will respond well to treatment in the beginning, between 60% and 90% of patients will relapse.
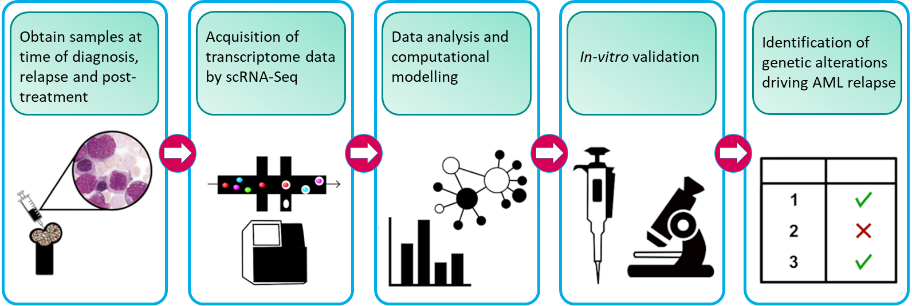
Our research project aims to understand what makes leukemic cells drug-resistant. By understanding how drug resistance occurs, we can develop strategies to prevent this from happening, improve the treatment, and prevent relapse. In order to do this, we are analysing individual cells from patients at various stages of their disease. We look at their cells when they are first diagnosed after they have had treatment, and then when the disease comes back. This approach allows us to identify sub-groups of AML cells in patients and find out which cells are sensitive to drugs and which ones are resistant.
From this, we can determine what happens inside individual AML cells that make them drug-resistant. In doing so we will be able to identify the group of AML cells and the mechanisms that are causing disease relapse. We also analyse healthy blood cells. This is important to find features that are only present in AML cells but not in healthy blood cells, otherwise targeting it for treatment will cause bad side effects. Mechanisms that are only active in AML cells are better targets.
Overall, results from our project are hoped to lead to new treatments able to overcome drug resistance and doing so prevent relapse and give a long-term cure to leukaemia patients.
Project Overview
Acute myeloid leukaemia (AML) is one of the few cancers where, despite the substantial increase in our understanding the gene defects driving the disease, the mortality rate is still increasing, with the overall 5-year survival being less than 20%. The main reason for this poor prognosis is drug resistance, which causes 60-90% of AML patients to relapse, at which stage there are no effective treatment options. Drug resistance is due to leukaemia cell heterogeneity and the bone marrow microenvironment (BMM) which protects and sustains the viability of the leukemic cells.
Here, we aim to tackle this unmet clinical need via a novel approach: understanding how the tumour microenvironment interacts and protects the malignant cells that initiate the disease, i.e. the leukemia stem cells (LSC). In this project, we are working towards identifying the genetic determinants LSC subpopulations that drive drug resistance and relapse, and computationally model how the BMM and LSC interaction underpin leukemogenesis, drug resistance and relapse after treatment.
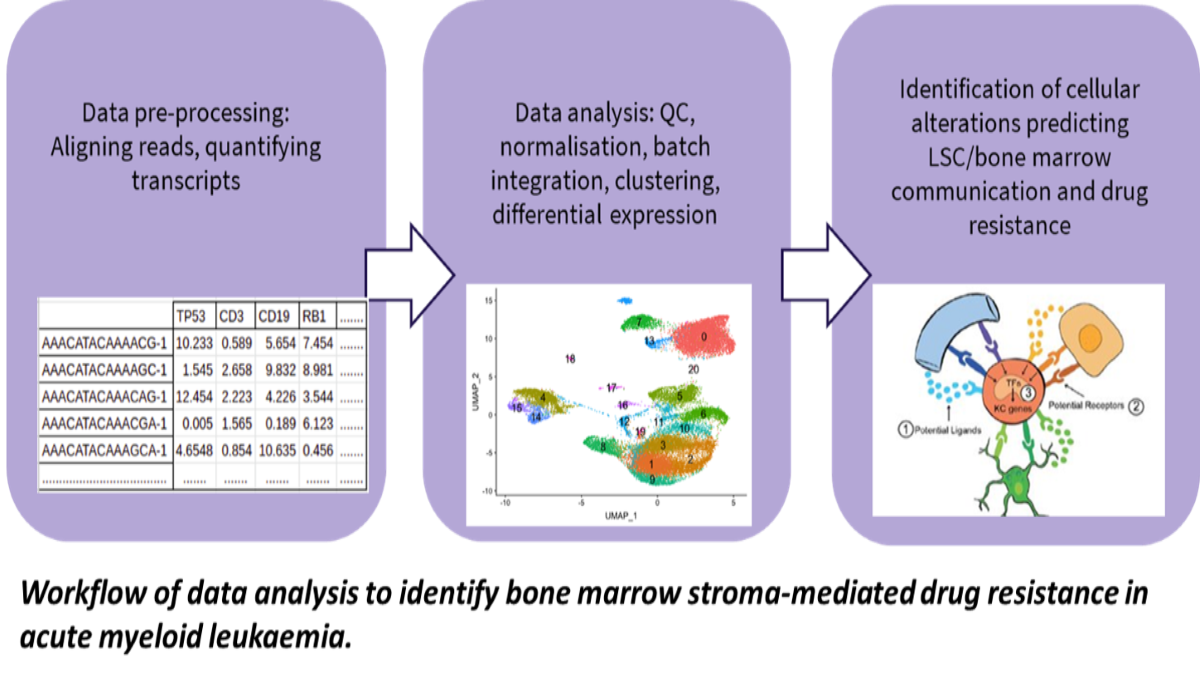
We have previously shown that some LSCs are resistant to clinically used chemotherapeutics and thus may drive relapse (O’Reilly et al., Sci Reports, 2018; Dhami et al., Br. J. Haematol., 2020). To determine which type of LSCs are resistant to drugs and why, AML patient samples are exposed to chemotherapeutics and the genetic make-up of the LSCs is determined using single cell transcriptomics. In parallel, LSC subpopulation heterogeneity in patient samples collected at diagnosis, after chemotherapy, and at relapse is determined using data from the Blood Cancer Biobank Ireland (BCBI) and the GEO public database (GSE66525, GSE35907). New computational methods for identifying distinct cell populations from mixtures are applied to identify the drug resistant subpopulations driving relapse. Analysis of the transcriptomics data using the computational modelling guides the design of new drug treatment combinations and their validation in ourex vivoculture models (Dhami et al., Drug Disc Today, 2016).
In order to understand the role of the BMM in AML, we use computational methods to determine cell type-specific genetic alterations and altered cellular composition of the BMM from genomic and genetic datasets from BM samples. With this data, we aim to develop a computational model of the AML-BMM system enabling the simulation of a patient’s response to treatments.

The team’s academic partner, Dr. Szegezdi, has expertise in AML biology, relevant AML and BMM culture models, and the industry team at Celgene are experts in advanced computational analysis methods.
For further information, visit
(opens in a new window)www.bloodcancers.ie
(opens in a new window)https://www.celgene.com/research-development/medical-innovation/
Key References:
- O' Reilly E, Dhami SPS, Baev DV, Ortutay C, Halpin-McCormick A, Morrell R, Santocanale C, Samali A, Quinn J, O'Dwyer ME, Szegezdi E. (2018) Repression of Mcl-1 expression by the CDC7/CDK9 inhibitor PHA-767491 overcomes bone marrow stroma-mediated drug resistance in AML. Sci Rep. 8: 15752; PMID: 30361682
- Dhami SPS, Tirincsi A, Baev D, Krawczyk J, Quinn J, Cahill MR, Zeugolis D, Szegezdi E. (2020) Theranostic drug test incorporating the bone-marrow microenvironment can predict the clinical response of acute myeloid leukaemia to chemotherapy. Br J Haematol. 189:e254-e258; PMID: 32342487.
- Dhami SPS, Kappala SS, Thompson A, Szegezdi E. (2016)(opens in a new window)Three-dimensional ex vivo co-culture models of the leukaemic bone marrow niche for functional drug testing.Drug Discov Today. 21: 1464-1471; PMID: 27130156
Project team