

P1.8: Whole genome and transcriptome sequencing of extreme phenotypes in gastrointestinal cancers
Plain Language Summary
Each individual patient’s cancer has an equally individual genetic profile. By studying this, scientists can find out valuable information to support both accurate diagnosis and optimal treatment.
So-called ‘extreme’ cases are those cancer patients who may have an unusually good or an unusually poor outcome, or those that respond extraordinarily well to treatment, or don’t respond at all. These patients can represent real-life models of a drug working perfectly as it should, or where a cancer can efficiently bypass the ‘block’ imposed by the drug. By focusing on these ‘extreme cases’ in our genetic studies, we can maximise the information that we get in each case. This allows scientists to get more information from fewer cases, reduces uncertainty by removing the ‘middle ground’, and increases the odds of making significant discoveries.
This project aims to identify patients with ‘extreme cases’ of gastrointestinal cancer, and analyse their genetic information. The aim is to identify the genetic factors causing these cancers, and find new ways to treat them more successfully. Our ultimate goal is to use this type of genetic study to support and guide clinical decisions for all cancer patients.
Visual Abstract
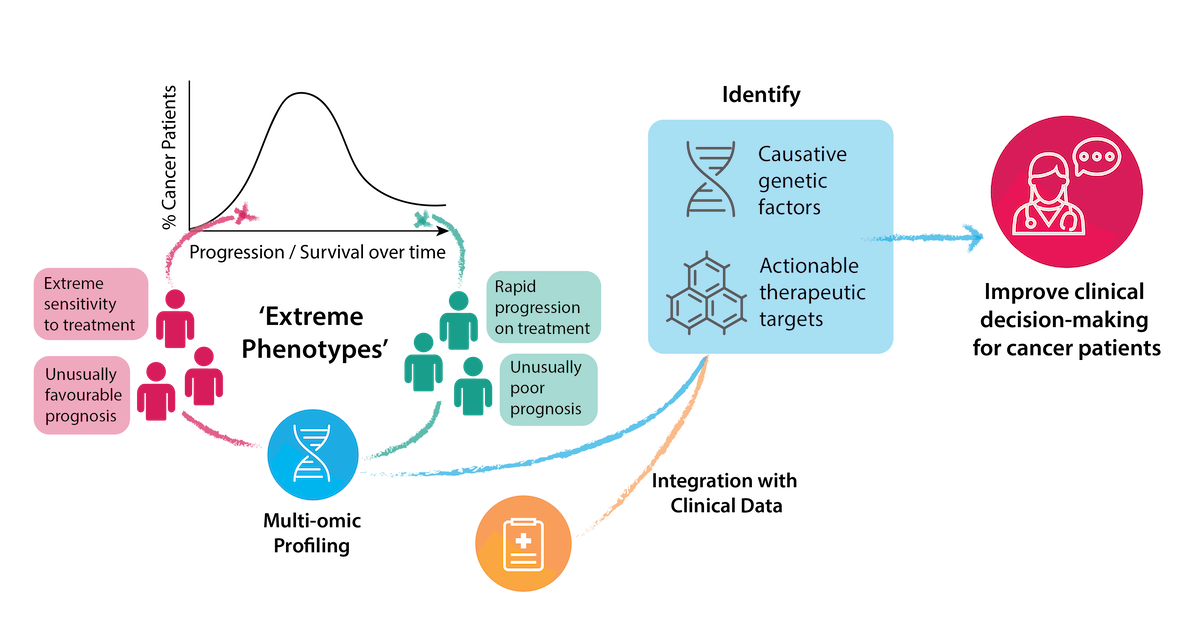
Project Overview
Personalised evaluation of the genetic signature of an individual cancer can provide both diagnostic information and enable optimal selection of therapy. However, the identification of clear predictive or prognostic biomarkers is challenging in a heterogeneous patient population. The identification and study of ‘extreme phenotype’ cancer patients can maximise the insights of genome sequencing, both in revealing the molecular causes that explain those phenotypes, and identifying useful therapeutic targets.
‘Extreme phenotype’ patients can be those with either an unusually favourable or an unusually poor prognosis, or a striking response or progression following treatment. Such patients represent real-life clinical models of drug sensitivity, resistance and toxicity, which facilitates the study of underlying molecular mechanisms. Extreme phenotype selection in cancer patients enhances the clinical interpretation of data generated by high-throughput techniques, and increases the likelihood of clinically significant discoveries. It also reduces the sample size required for informative comprehensive genome profiling, by natural enrichment of biomarker expression in patients studied, and exclusion of confounding intermediate phenotypes of uncertain significance.
This project aims to identify patients with extreme GI cancer phenotypes, and perform whole genome / transcriptome sequencing on tumour and germline samples from patients. By integrating clinical information with the results of these genetic analyses, we can identify potential causative genetic factors and actionable therapeutic targets in these cancers. Ultimately, we are aiming to integrate genome and gene expression based clinical decision-making into the clinical routine for treating cancer patients.
The project is a strategic collaboration between Trinity St James Cancer Institute (TSJCI), Genuity Science, and the British Columbia Cancer Agency (BC-CA), Canada, represented by Prof Janessa Laskin. Our collaborators at the BC-CA are leading a large Personalized OncoGenomics study since 2012, and have analysed over 1000 samples from patients with advanced refractory cancer through whole genome / transcriptome sequencing. Clinical and genetic data are presented at a multidisciplinary molecular tumour board, where clinical implications of individual patient results are reviewed and discussed. We have adopted this innovative approach and implemented it in Dublin. The overall goal is to build capacity for the real time comprehensive genetic analysis of patients with extreme cancer phenotypes in Ireland, and evaluate the therapeutic implications. The National Cancer Strategy 2017-2026 clearly emphasises the need for an improvement in cancer genetics and cancer molecular diagnostic services in Ireland. Currently, clinical genetic sequencing for cancer, both germline and somatic, is limited to screens of known cancer genes either as commercial panels or at academic centres. The urgently needed capacity to functionally interpret whole genome/transcriptome sequencing of tumour biopsy samples with real time clinical and therapeutic implications does not currently exist in Ireland.
Project team



