

P1.5: Assessment of genomic and proteomic anomalies in breast cancer
Plain Language Summary
In approximately 30% of breast cancer patients, the disease spreads from the initial primary tumour site to other parts of the body, known as metastasis. This process is not well understood, and it is difficult to predict in advance which cancers will metastasise.
This project is examining the genetic profiles of matched primary and metastatic tumour samples, using cutting-edge laboratory and computational techniques. By comparing the profiles of these matched samples, we are aiming to identify a panel of genes, or ‘biomarkers’ that indicate a high risk of metastasis.
Our aim is that these biomarkers can then be assessed at the point of diagnosis, and used to predict which patients are at a high risk of metastasis. These patients may then be offered more aggressive treatment to reduce their risk. In the future we hope that some of the biomarkers identified may become targets for new therapies to specifically inhibit breast cancer metastasis.
Considering that more than half of the patients diagnosed with breast cancer metastases have limited treatment options and typically succumb to disease within a few years, this addresses an unmet clinical need.
Visual Abstract
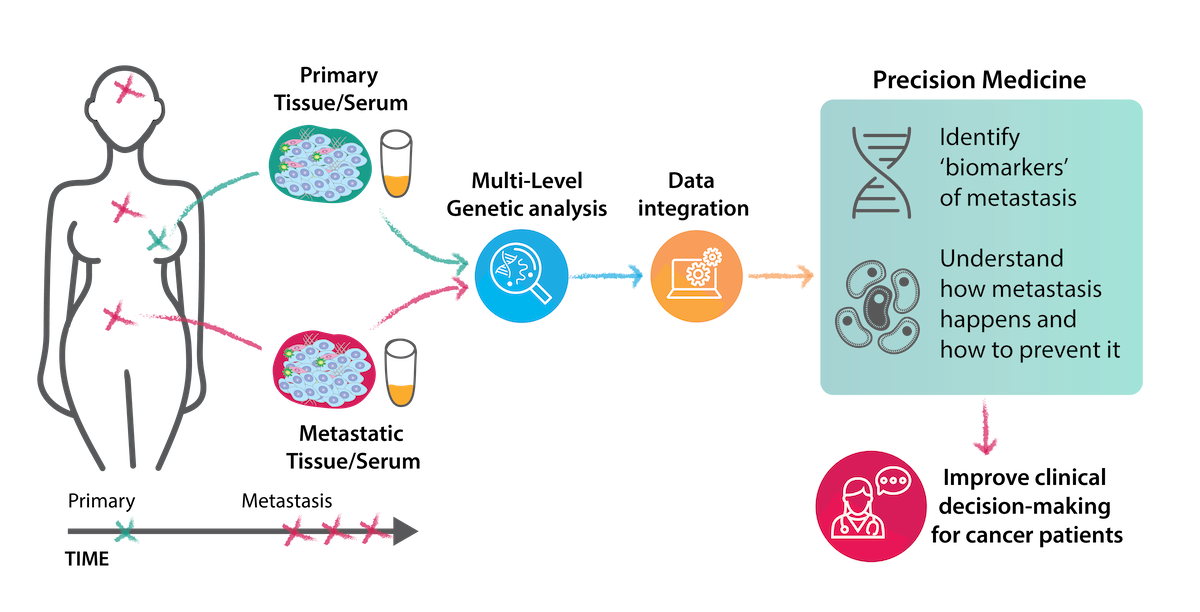
Project Overview
Metastasis occurs in approximately 30% of breast cancer patients, but the mechanism of progression to metastatic disease remains largely unknown, and thus far there are no predictive markers of disease progression. This project is applying cutting-edge techniques to identify a panel of biomarkers predicting metastatic progression in breast cancer. Considering that more than half of the patients diagnosed with breast cancer metastases have limited treatment options and typically succumb to disease within a few years, this addresses an unmet clinical need.
This project is applying a multi-layered genomic and epigenomic profiling approach on metastatic tumour material. Matched primary and metastatic tumours are profiled utilising exome, transcriptome and methylation sequencing. To analyse and integrate these data, we have established a comprehensive computational pipeline, taking advantage of the expertise in the POI consortium. Multi-omics profiling and data integration is now seen as the way forward in precision medicine. This project is pioneering this approach using a well annotated patient cohort to tackle a clinically important problem in breast cancer.
Our group previously characterized the metastatic-altered transcriptome across 21 patient-matched primary breast tumours and their associated metastases, identifying key adaptive pathways. We observed an enrichment of druggable kinase-driven signalling (RET, ERBB2) and activation of the HER2 pathway in metastasis. In contrast, several clinically actionable genes and tumour suppressors were down-regulated in metastasis relative to the primary tumour. For instance, the oestrogen receptor α transcript, ESR1, was consistently depleted in metastases, which correlated with hormone therapy resistance, and with increases in HER2 signature. Further, we observed a gain in global methylation in patient metastatic tumours in comparison to matched primary tumour tissue. Treatment with epigenetic modulators had anti-tumour activity ex vivo and could induce the re-expression of differentiation-promoting, tumour suppressor genes. Thus, we hypothesise that recurrent transcriptional remodelling, which is in part epigenetically regulated, is central to the development of metastatic disease.
Building on these results we are performing multi-omics profiling of 30 primary and metastatic tumour samples, both tissue and serum, using next-generation exome sequencing, RNAseq and Methylseq. The results of this profiling will be integrated with clinical data with the aims to (i) identify novel biomarkers; and (ii) to determine which combinations of datasets are the most informative. As these biomarkers represent control nodes in the disease network, they also may lend themselves as therapeutic targets.
Project team



